Overview
This building block provides you with some practical tips for dealing with large datasets in R.
Many R users rely on the base R commands read.csv or read.table to import their datasets as a dataframe. Although this works well for relatively small datasets, we recommend using one of the following packages instead: the readr, data.table or the vroom package - simply because it is significantly faster and offers enhanced functionality.
You can obtain the data for the examples from this link.
Advantages of Using readr, data.table, or vroom Over Base R
When working with large datasets in R, the default read.csv method can be less efficient in terms of speed and memory usage. This is where packages like readr, data.table, and vroom come into play, offering a more optimized approach to data import.
1. Significant Speed Gains
Efficiency is important when processing large datasets in R. Traditional base R methods, while reliable, do not offer the speed necessary for modern data analysis tasks. This is where readr, data.table, and vroom shine, each providing solutions to significantly reduce data loading times.
Practical Impact
Benchmarking reveal that all three packages significantly outperform the base R function in terms of speed. Specifically, these packages can load the dataset, with 1,809,903 rows and 12 columns, from 3 times up to 11 times faster!
# Load required libraries
library(readr)
library(data.table)
library(vroom)
# Time comparisons for reading a large file
system.time(base_r <- read.delim("GB_full.txt"))
system.time(readr_data <- read_tsv("GB_full.txt"))
system.time(dt_data <- fread("GB_full.txt"))
system.time(vroom_data <- vroom("GB_full.txt", col_names = FALSE))
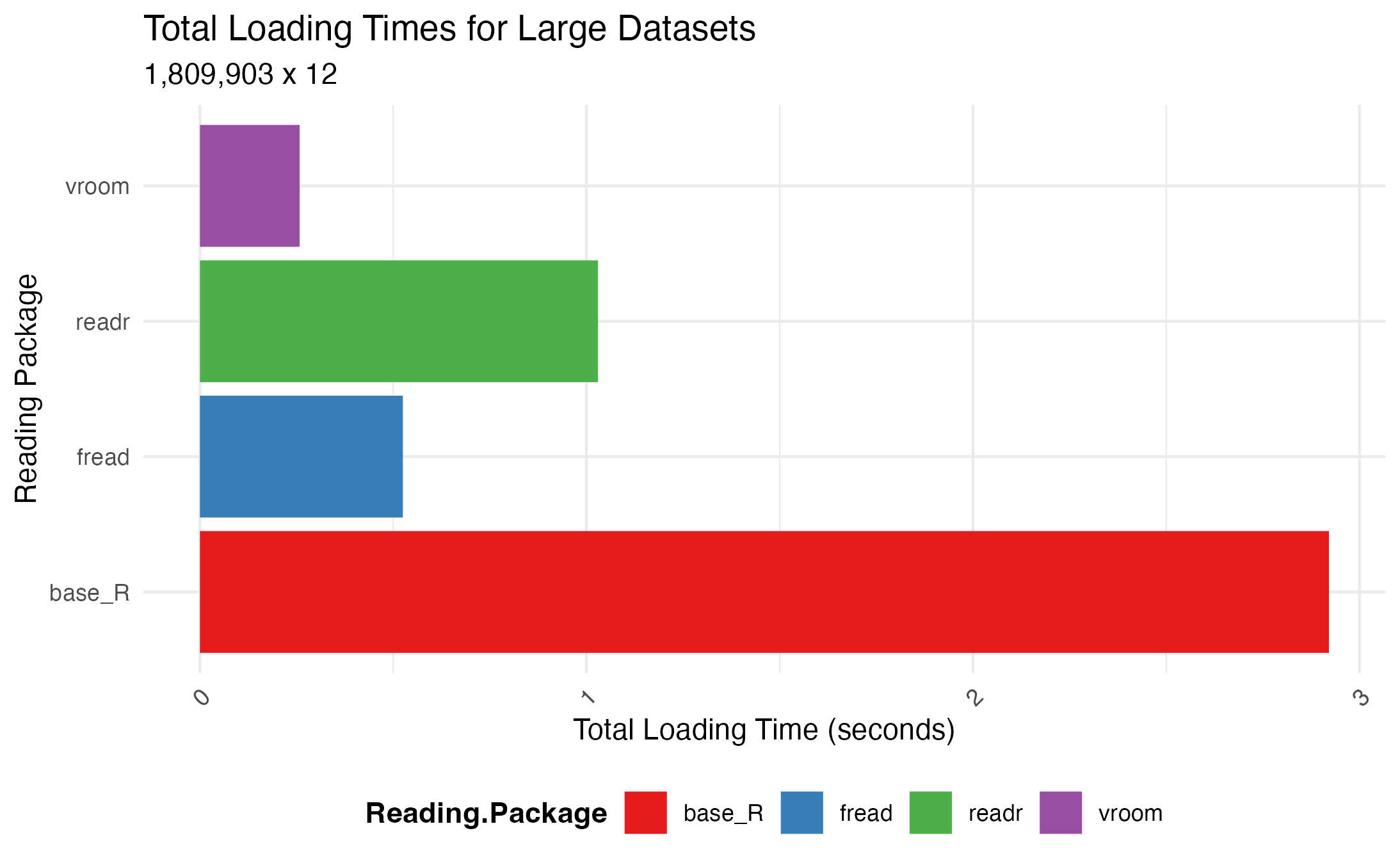
2. Quick Data Manipulation
The ability to efficiently manipulate data is as crucial as the import phase. All three packages readr, data.table, and vroom are coming shipped with data manipulation features to improve convienence, speed and memory usage.
Readr Package:
- Selective Loading:
readrprocesses the data while loaded, not after, facilitating faster manipulation. - Automatic Typing: Automatically infers column types, reducing the need for post-import adjustments and speeding up the data preparation process.
Data.table Package:
- In-Place Modification:
data.tableallows for in-place data modification, avoiding the slowdowns caused by copying data. - Keys and Indices: Setting keys for data indexing accelerates sorting and subsetting operations.
- Multithreading: Utilizes all CPU cores for parallel processing of tasks.
- Minimal Copying: Reduces memory usage by avoiding data duplication.
Vroom Package:
- Initial Load: Uses a quick scan to map data locations, reducing initial load times.
- Lazy Loading: When filtering and aggregating
vroomonly loads in the only necessary data, increasing speed performance and reducing memory footprint. - Capability for Large Datasets: By querying and subsetting of datasets, avoiding full data materialization, it allows for datasets larger than the memory.
Practical impact
In the benchmarks, we explore different data manipulation tasks like printing, head/tail operations, random row sampling, specific row filtering, and aggregating for mean calculations.
Using the following methodology:
- vroom_base: Uses
vroomfor reading, thenbase Rfunctions for manipulation. - vroom_dplyr: Integrates
vroomfor reading withdplyrfor manipulation. - data.table: Employs
freadfor reading anddata.tablefunctions for manipulation. - readr_dplyr: Uses
readrfor reading anddplyrfor manipulation. - base_R: Relies on standard
base Rfunctions throughout.
The table below represents the running times observed for different data manipulation packages:

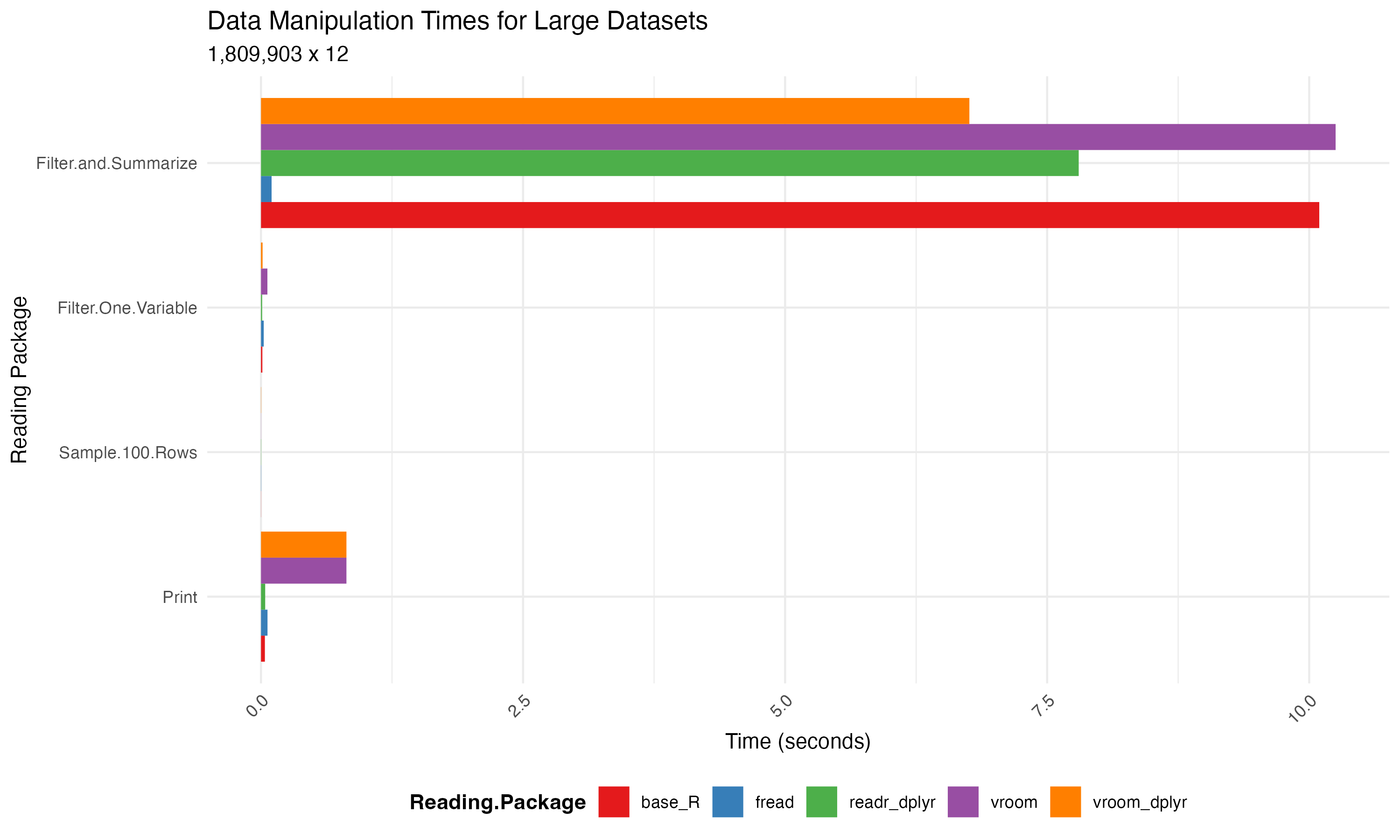
3. Increases File Writing Performance
When it comes to writing large datasets to files, readr, data.table, and vroom offer increased speed performance compared to base R’s write.csv() function.
Here is our benmark test to compare these functions:
# creating a 1 million by 10 data frame
df <- data.frame(matrix(rnorm(1000000*10), 1000000,10))
system.time({write.csv(df, "base.csv", row.names=FALSE) }) # base R
system.time({fwrite(df, "datatable.csv") }) # data.table
system.time({write_csv(df, "readr.csv") }) # readr
system.time({vroom_write(df, "vroom.csv")}) # vroom

Below is a visual representation of the file writing times across different packages:
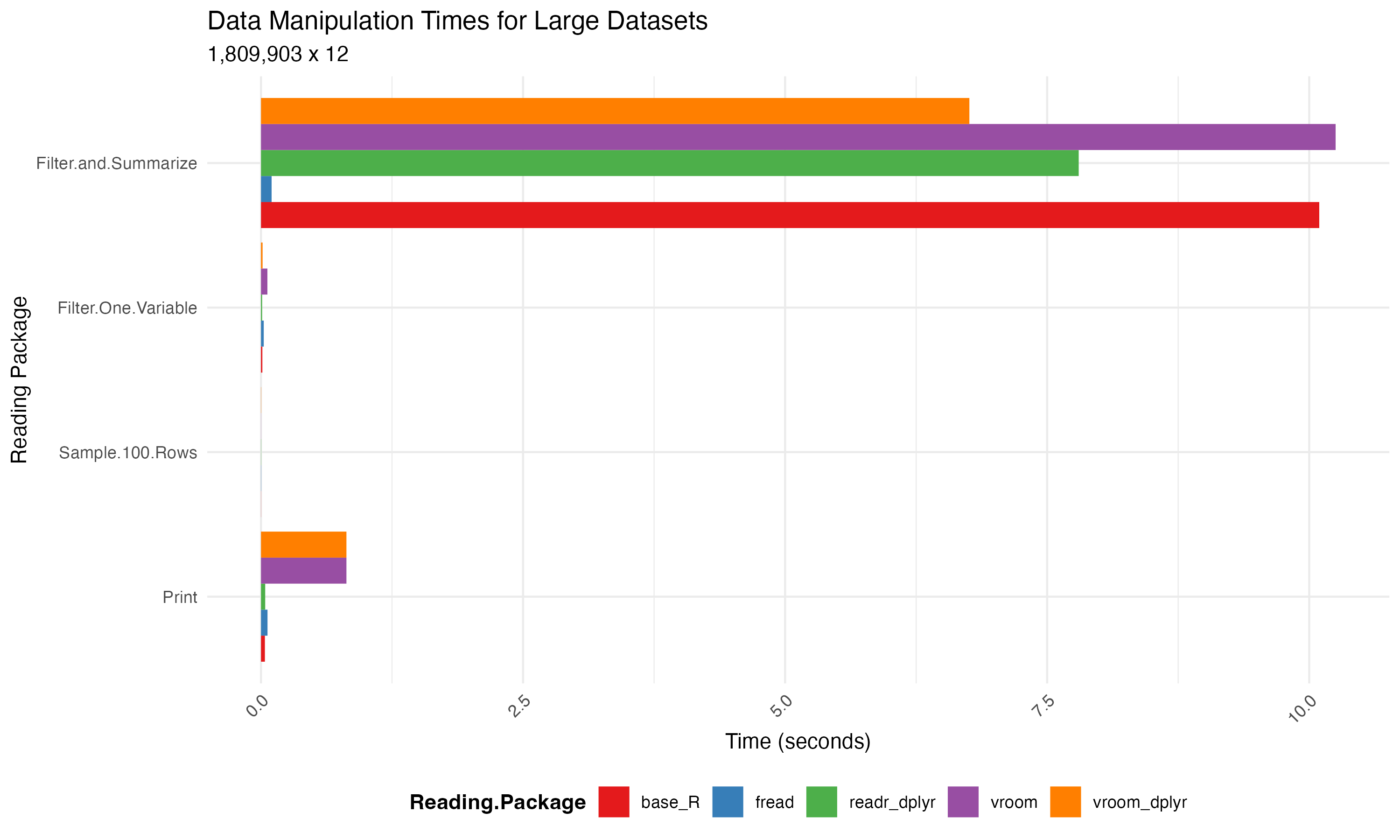
Practical Examples
This section walks you through practical examples using the various R packages for data importation of large datasets.
Importing with Readr
read_csv() from the readr package offers several advantages over the base R function read.csv():
- Integration and Type Detection:
read_csv()works well with othertidyversepackages and automatically determines the data type for each variable. - Tibble Output: Unlike
read.csv(), which produces a data.frame,read_csv()outputs a tibble. Tibbles offer enhanced functionality and behave more consistently when subsetting. - Character Variable Handling:
read_csv()treats text variables as character variables.- In contrast,
read.csv()automatically converts text to factors, which can be inconvenient.
- In contrast,
Overall, read_csv() provides a more efficient and user-friendly approach to data handling in R.
# import package
library(readr)
# Import data with the readr package
df <- read_csv("YOUR_DATASET.csv")
# Import a maximum number of rows for exploratory analysis
df <- read_csv("YOUR_DATASET.csv", n_max = 500)
# Import data that you want to use
df <- read_csv(("YOUR_DATASET.csv"), col_select = c(1, 3:4)) # column indices
df <- read_csv("YOUR_DATASET.csv", col_select = c("date","country","revenue")) # column names
# store the derivative file for future use
write_csv(df, "YOUR_CLEANED_DATASET.csv")
 Tip
TipReadr comes with several other functions. Such as read_csv2() for European-friendly variant comma-separated data, read_tsv() for tab-separated data, read_lines() for line-by-line file extraction and read.txt() reading fixed width data.
Importing with data.table
Switching from the read.csv() function to fread() can improve the performance of your program. It is often dubbed the “fast and friendly file finagler”, and is efficient and straightforward to use in R.
One of its main features is that it is designed to import data from regularly delimited files directly into R. Here, “regular” implies that each row in your dataset must have the same number of columns. What sets fread() apart is its ability to automatically detect parameters like sep, colClasses, and nrows, making the data import process straightforward.
The code block below illustrates how you can import your whole dataset using fread() as well as subsets of it, determine the size of the resulting object, and store your new/clean versions of the data as a new file for future use.
# import package
library(data.table)
# import data with data.table package
df <- fread("YOUR_DATASET.csv")
# only import the first couple of rows for exploratory analysis
df <- fread("YOUR_DATASET.csv", nrows=500)
# only import the data you actually use
df <- fread("YOUR_DATASET.csv", select=c(1, 2, 5)) # column indices
df <- fread("YOUR_DATASET.csv", select=c("date", "country", "revenue")) # column names
# print object size in bytes (for a quick comparison)
object.size(df)
# store the derivative file for future use
fwrite(df, "YOUR_CLEANED_DATASET.csv")
 Tip
TipWhen starting your analysis with large datasets, start with loading a representative sample first. This practical strategy facilitates quicker fine tuning and testing of data processing and analysis scripts. If confident about hte accuarcy, and robustness of your code, scale up to the entire dataset. This approach saves time upfront and ensures a smoother workflow when handling extensive datasets.
Importing with vroom
Vroom is part of tidyverse and is a similar to readr::read_csv() or data.table::fread(). However, when it comes to raw importing speed performance, vroom::vroom() often leads the pack. In the context of the dataset examined within this article, it processes data in approximately one-quarter the time taken by data.table::fread().
The efficiency of vroom is attributed to its method of reading in files. Initially, it only marks the location of each data point, deferring from actual reading of values. Thereafter, it uses, the so-called Altrep framework, which creates vectors that reference these marked locations. This two-step procedure makes data acces fast and memory efficient.
Therefore the main reason vroom is faster is because character data is read from the file lazily; it does not read the entire dataset unless required. One major benefit is that it requires no adjustments to your data-manipulation code. However, one disadvantage to vroom’s lazy reading method is that it may delay the identification of potential data issues until the point of access.
Observations from benchmark results show that vroom’s initial read is significantly quicker than alternative methods. Routine operations such as print(), head(), and random sampling execute equally fast as the other packages. However because the character data is read lazily, operations such as filter and summarise, which need character values, require additional time. However, this cost will only occur once. After the values have been read, they will be stored in memory, and subsequent accesses will be as quick as the other packages.
# import package
library(vroom)
# get path to example file
input_file <- vroom_example("YOUR_DATASET.csv")
input_file
# import data with the vroom package
# Read from a path
df <- vroom(input_file)
# You can also use your path directly
df <- vroom("YOUR_DATASET.csv")
# only import the first couple of rows for exploratory analysis
df <- vroom("YOUR_DATASET.csv", nrows=500)
# only import the data you actually use
df <- vroom("YOUR_DATASET.csv", col_select=c(1, 2, 5)) # column indices
df <- vroom("YOUR_DATASET.csv", col_select=c(date, country, revenue)) # column names
# store the derivative file for future use
vroom_write(df, "YOUR_CLEANED_DATASET.tsv")
vroom_write(df, "YOUR_CLEANED_DATASET.csv", delim = ",")
 Tip
TipOftentimes, datasets you previously worked with remain stored in memory, even if they’re no longer in use. In RStudio, click on the broom icon in the top right window to remove all objects from the environment. By removing objects which are no longer in use you will help improve RStudio’s performance and reduce the risk of errors and conflicts.
 Summary
SummaryThis article discusses efficient strategies for handling large datasets in R using the readr, data.table, and vroom packages. Key takeaways are:
readr,data.table, andvroomare faster for data importation, data manipulation and writing files compared to base R functions.readroffers advantages like selective loading and automatic typing, improving the speed of data manipulation.data.tablesfread()has the ability to automatically detect parameters like sep, colClasses, and nrows, making the data import process straightforward.vroomaccelerates initial loads through quick scans and lazy loading, supporting efficient querying and subsetting of large datasets.- Benchmark comparisons demonstrate the largely improved performance of these packages in both data import and manipulation tasks.
- Practical examples guide you through using each package for importing, exploring, and manipulating large datasets.





